Intro to R Notebooks
This notebook is based loosely on this cheatsheet that shows some of the most basic functionality of R notebooks, including code, markdown, and Latex.
Code
Chunks
You can imbed chunks of R code right into the notebooks and run them. Why not beat that long-dead horse of demo data, the iris dataset?
head(iris)## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosaIf you print a dataframe, it automatically gets formatted in this nice way.
We can also look at the summary:
summary(iris)## Sepal.Length Sepal.Width Petal.Length Petal.Width
## Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100
## 1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300
## Median :5.800 Median :3.000 Median :4.350 Median :1.300
## Mean :5.843 Mean :3.057 Mean :3.758 Mean :1.199
## 3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800
## Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500
## Species
## setosa :50
## versicolor:50
## virginica :50
##
##
## And of course, you can use whatever libraries you like, as usual. We can set an option in this chunk to disable the messages that print to the console when you do things like load libraries. We could also turn off warnings, set options for error handling, or specify how we want results and code interleaved, or if we want the code in the output document at all. Setting the tidy argument to TRUE also spiffs up your code for you (cleaning up multiline expressions with indenting, for instance) in the output. So customizable!
library(dplyr)
(meanpetals <- group_by(iris, Species) %>%
summarize(length = mean(Petal.Length), width = mean(Petal.Width)))## # A tibble: 3 x 3
## Species length width
## <fct> <dbl> <dbl>
## 1 setosa 1.46 0.246
## 2 versicolor 4.26 1.33
## 3 virginica 5.55 2.03Boring old data demos never looked so fresh!
Inline
Inline code executes invisibly, so all you see is the output. For example, I can run a function right here and now to get the weekday:
Today is a Monday.
That was accomplished by simply adding a back-ticked r prefix to a call to weekdays(Sys.time()).
You can also set global parameters in the header and reference them throughout the document.
This inline execution has various uses. If you need code primarily for its output, like getting the current date or weekday and adding it into text, then inline works really well and better than a chunk.
Displaying Data
If you’re writing up a tutorial or a report on results, you’ll more likely than not need to display data in the form of plots and tables. This is super easy in R Notebooks, and there are lots of options.
Plots
Let’s see what the sepal lengths in Iris are up to by species, just for kicks. I’ll do some more extensive data manipulation in a code chunk, just to show we can. Since I loaded dplyr in another code chunk further up, we’re good there.
library(ggplot2)
iris_plot <- ggplot(data=select(iris, Species, Sepal.Length),
aes(x=Species, y=Sepal.Length, color=Species, fill=Species)) +
geom_violin(alpha=.1) +
geom_point(position=position_jitter(w=.2)) +
geom_crossbar(stat='summary',fun.y=mean, fun.ymax=mean, fun.ymin=mean, fatten=2, width=.5) +
theme_minimal()
print(iris_plot)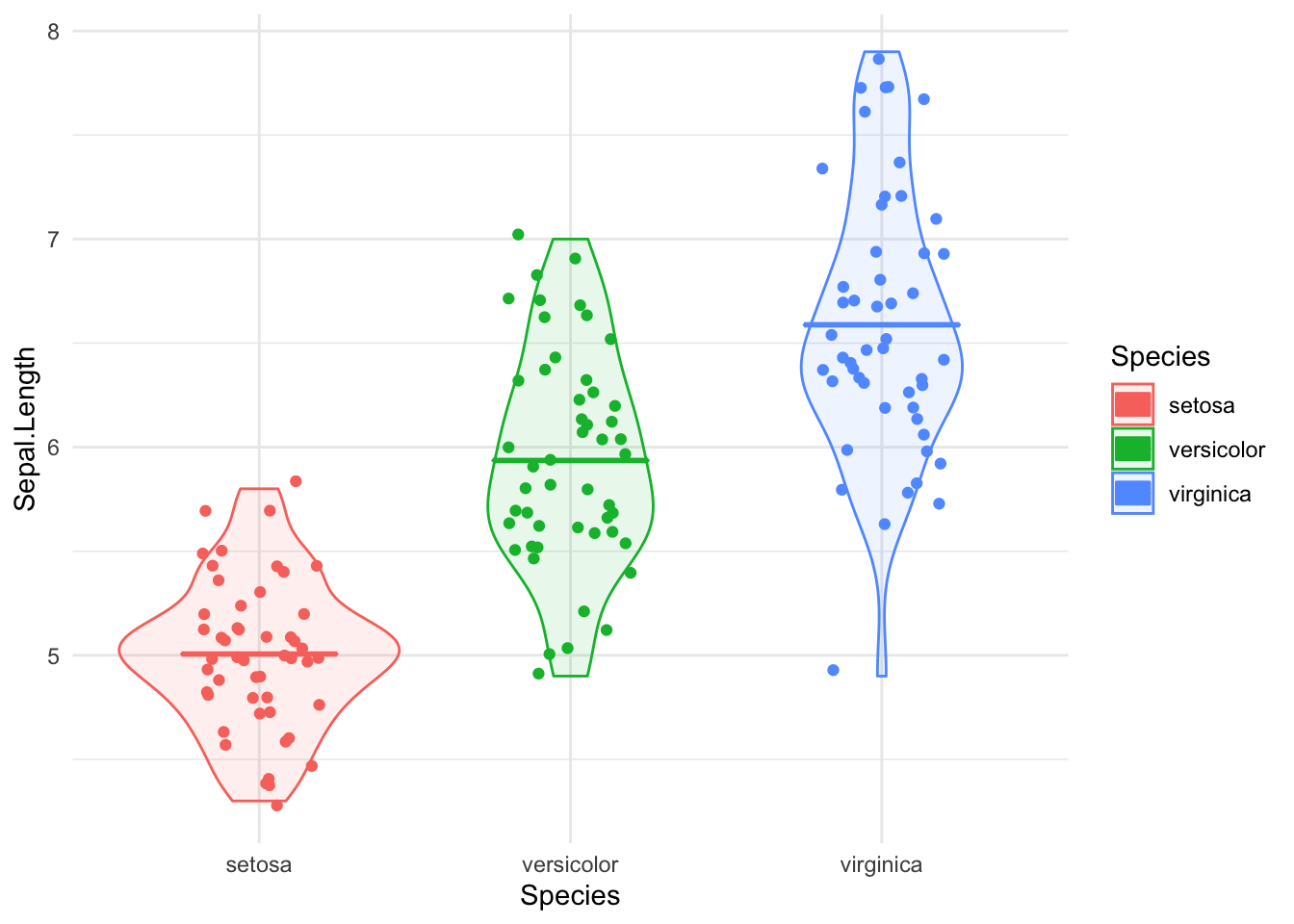
Fascinating, I guess!
The nice thing about this is that these plots are completely reproduceable–they’ll be generated everytime you run the notebook. You can keep the code cell that generates it in the output, so people can see it, or if you want a cleaner output with just the results, you can hide the generating code cell. You can tailor it to your options.
Tables
Tables can be a real pain-point when doing manuscripts, but these can be automated in R Notebooks, rendered right into text, and some look pretty good. Here’s what knitr will give you (it won’t look right until it’s rendered):
knitr::kable(meanpetals)| Species | length | width |
|---|---|---|
| setosa | 1.462 | 0.246 |
| versicolor | 4.260 | 1.326 |
| virginica | 5.552 | 2.026 |
That looks alright!
There’s also xtable, which will make it easier to set the appearance of the table through HTML attributes. This won’t show up until it’s rendered, though.
print(xtable::xtable(meanpetals), type='html', html.table.attributes='border=0 width=250')| Species | length | width | |
|---|---|---|---|
| 1 | setosa | 1.46 | 0.25 |
| 2 | versicolor | 4.26 | 1.33 |
| 3 | virginica | 5.55 | 2.03 |
Finally, here’s a table fron the stargazer package. Also lots of attributes we can set here. It’s designed mostly for pretty formatting of regression model results and summary stats, but we can get it to do direct output instead.
stargazer::stargazer(as.data.frame(meanpetals), type='html', summary=F)| Species | length | width | |
| 1 | setosa | 1.462 | 0.246 |
| 2 | versicolor | 4.260 | 1.326 |
| 3 | virginica | 5.552 | 2.026 |
If none of the formatting is quite to your liking, or you don’t have easily available options to set, you can tweak the appearance with your own CSS.
If none of this is satisfying, or it’s more informal, you can always just print the dataframe.
(meanpetals)## # A tibble: 3 x 3
## Species length width
## <fct> <dbl> <dbl>
## 1 setosa 1.46 0.246
## 2 versicolor 4.26 1.33
## 3 virginica 5.55 2.03I don’t know that this makes tables any less painless, but at least now there’s more than one option for the pain.
Formatting
If you’re writing up a report or manuscript, you also need your text to look nice, not just your data. This is where knowing a little bit of Markdown comes in handy.
Markdown
Just type to get pretty plaintext. To quote verbatim code without running anything, wrap it in backticks(`). This is distinct from inline code, which is prefaced with r within the ticks. I’ll do this throughout to demonstrate how to construct the effects you see.
To display a special character, like _underscores_ or asterisks *, escape them with a backslash (\).
To add a single linebreak without a blank line, end a line with two spaces...
Ta-da!
*italics*: italics
**bold**: bold
**_both_**: both
~~strikethrough~~: strikethrough
subscript~1~: subscript1
superscript^2^: superscript2
You can do bulleted lists:
* start with an asterisk, plus, or minus + then indent 4 spaces - then indent again * then more stuff
- start with an asterisk, plus, or minus
- then indent 4 spaces
- then indent again
- then indent 4 spaces
- then more stuff
And ordered ones:
1. Big thing i) small thing \+ indent A. sub-small thing 2. Another big thing
- Big thing
- small thing + indent
A. sub-small thing
- small thing + indent
- Another big thing
Oh, and footnotes… [^1] 1
#Header 1
Header 1
##Header 2
Header 2
###Header 3
Header 3
Add horizontal lines with at least three hyphens, asterisks, or underscores:
***
Tables are a little strange in the raw, but look nice once rendered.
| Right-aligned | Left-aligned | Centered | Default |
|-:|:-|:-:|-|
|1|1|1|1|
|12|12|12|12|
|*1*|_2_|~~3~~|4^2^|| Right-aligned | Left-aligned | Centered | Default |
|---|---|---|---|
| 1 | 1 | 1 | 1 |
| 12 | 12 | 12 | 12 |
| 1 | 2 | 42 |
Images are easy, too.


The adorabilis octopus, for your viewing pleasure
[^1]:Are pretty easy.↩
